p53 – Guardian of Human Genome
April 20, 2023 2023-04-25 6:21p53 – Guardian of Human Genome
Do you know 50% of cancers are caused by innate genes when mutations inactivate them? The best example of this is the Tumor Suppressor Genes. They are innate genes that suppress tumors by slowing cell division, repairing damaged DNA, and regulating cell apoptosis.
During normal development, cells are designated for programmed cell death or apoptosis. If apoptosis fails and the cells keep proliferating, it may lead to tumor formation, as in the case of Chronic Lymphoblastic Leukemia.
You must be thinking, why we are discussing all these without making any point about p53? Well, here comes an answer to that.
p53 is a protein located on the 17th chromosome and the most common pro-apoptotic tumor suppressor gene. When the function of p53 is lost, apoptosis cannot be induced, and the accumulation of mutations required to develop cancer becomes more likely.
Despite its unassuming name, the p53 gene may have more to do with the development of human cancer than any other genome component. p53, when absent, is responsible for a rare inherited disorder called Li-Fraumeni Syndrome. Victims of this disease are afflicted with a very high incidence of various cancers, including breast, and brain cancer and leukemia. Tumors of cells bearing p53 mutations correlate with much less survival rate than those containing a wild-type p53 gene.
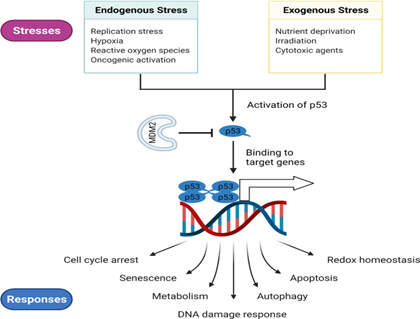
How exactly does this p53 gene works?
To understand the working of the p53 gene, we should first know about cell cycle checkpoints. Normal cells move in a cell cycle in a regulated way that makes sure that cells do not divide in unfavorable conditions. These cell cycle checkpoints are present whenever there is a transition from one phase of the cycle to another. The most important checkpoints are the G1 checkpoint (G1 phase to S phase transition), G2 checkpoint (G2 phase to M phase transition), and Spindle check point (during the transition of metaphase to anaphase).
The p53 protein is a sensor essential for checkpoint control that arrests cells with damaged DNA in G1. At the same time cells lacking available p53 cannot be arrested. Unlike other cell-cycle proteins, p53 is present at deficient levels in normal cells because of its extremely unstable and rapidly degraded.
Expression of the p53 gene is heightened only in stressful situations such as hypoxia, heat, and UV, gamma-radiations. DNA damage by all these leads to the activation of ATM, a serine kinase pathway that stabilizes p53, leading to a marked increase in its concentration. The stabilized p53 activates the expression of gene encoding p21, which binds to and inhibits mammalian G1 cyclin CDK complexes. As a result, to allow time for DNA repair, cells are arrested in G1.
If the repair is successful, the p53 and p21 levels will fall, and the cells can then progress into the S-phase. When the p53 G1 checkpoint control doesn’t operate properly, damaged DNA can replicate, perpetuating mutations and DNA rearrangement passed on to daughter cells, contributing to the likelihood of transformation into metastatic cells.
The activity of p53 is typically kept low by a protein called Mdm2, a negative regulator of the tumor suppressor p53, and plays a crucial role in controlling its protein stability. Mdm2 inhibits the transcription-activating ability of p53 by binding to it thus preventing excess p53 function.
The Mdm2 gene is amplified in many sarcomas and other human tumors that contain a normal p53 gene. Even though functional p53 is produced by such tumor cells, the elevated Mdm2 levels reduce p53 concentration enough to abolish the p53-induced G1 arrest.
Even though twenty thousand research papers on p53 have been released since 1979, no conclusive methods for detecting p53 are available even after thorough studies. So rather than targeting p53 directly, exploiting mutant p53 synthetic lethal genes might provide additional therapeutic benefits.
Authored by Vaishnavi, BITS Biocon Certificate Program in Applied Industrial Microbiology, Batch 9
References:
- Karp- Cell and Molecular Biology, 6th edition
- Lodish-Molecular cell biology, 5th edition
- Cooper- Cell biology
- https://doi.org/10.1186/s13045-021-01169-0








